LobeFinder
Purdue University
Overview
| Measured variables | shape, growth, surface |
|---|---|
| Operating system | windows, mac, linux, matlab |
| Licence | freeware |
| Automation level | semi-automated |
| Plant requirements | any |
| Export formats | csv |
| Other information | requires MatLab |
Scientific article(s)
LobeFinder: a convex hull-based method for quantitative boundary analyses of lobed plant cellsTzu-Ching Wu,Samuel Belteton,Jessica Pack,Daniel B. Szymanski,David UmulisPlant Physiology, 2016 View paper
Similar tools
Other tools for the analysis of cell,leaf:
Description
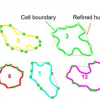
LobeFinder is a convex-hull based algorithm to identify lobes, quantify geometric properties, and create a useful graphical output for further analysis. The algorithm was validated against manually curated cell images of pavement cells of widely varying sizes and shapes. The ability to objectively count and detect new lobe initiation events provides an improved quantitative framework to analyze mutant phenotypes, detect symmetry-breaking events in time-lapse image data, and quantify the time-dependent correlation between cell shape change and intracellular factors that may play a role in the morphogenesis process.
Find more resources on https://github.com/George-wu509/LobeFinder
Modified from Plant Physiology article